Introduction to Immune Repertoire Sequencing

Introduction to Immune Repertoire Sequencing
What is immune repertoire sequencing?
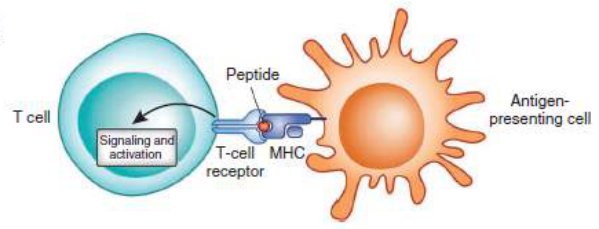
Figure1. Immune repertoire sequencing
The immune repertoire (IR) refers to the sum of all functionally diverse T and B cells within an individual's circulatory system at a certain time. The adaptive immune system of the human body relies mainly on the recognition and specific binding of MHC antigen peptide molecules by T cell receptor (TCR) and complementary determining region (CDR) on the surface of T and B lymphocytes and B cell receptor (BCR). Due to the limited human genome, it is impossible to encode a sufficient number of receptor genes to produce immune protection against various antigens in the outside world, but the VDJ gene encoding the CDR region of TCR on the surface of T cells has been rearranged, and B cells undergo VDJ gene rearrangement, high frequency somatic mutation, antibody class conversion after receiving antigen stimulation, forming the diversity of TCR and BCR. The body has the structural basis of immunity to a variety of foreign antigens; on the other hand, it also poses a great challenge to the systematic and comprehensive analysis of TCR and BCR libraries.
Immune repertoire sequencing is based on T lymphocytes and B lymphocytes. Multiplex polymerase chain reaction (PCR) or 5'RACE technology is used to amplify CDR that determines the diversity of TCR or BCR, combined with high-throughput sequencing technology and data analysis. Thus, the diversity of the immune system is assessed and the relationship between the immune repertoire and disease is studied in depth.
Development of immune repertoire sequencing technology
In the 1990s, immunoscan profiling was used to study BCR and TCR. Sanger sequencing is to sequence the DNA fragments encoding the variable region of BCR or TCR, but the number of sequencing is relatively limited, which restricts the application and promotion of this technology. Flow immunoassay technology and CDR3 scanning analysis technology can separate B cells or T cells, thus making up for the shortage of Sanger sequencing in quantity. Subsequently, through limited dilution method, single B cell antibody gene was cloned and expressed to study the characteristics and functions of specific antibodies, and then isolate disease-related antibodies, especially pathogen-related antibodies. Compared with Sanger sequencing technology, the second-generation high-throughput sequencing technology is currently the most widely used, which mainly relies on DNA template PCR amplification to enhance the fluorescence capture signal. Using the second-generation high-throughput immune repertoire sequencing technology, it can obtain unpredictable depth, resolution and accuracy, but the second-generation sequencing technology requires reverse transcription and PCR amplification. The subsequent three-generation sequencing methods do not require PCR amplification and directly detect a single base signal. Compared with the second-generation sequencing technology, the third-generation sequencing technology has higher accuracy. Second-generation high-throughput sequencing technology has high sensitivity and specificity, and monitors the expression of multiple cloned gene sequences at the same time, which is the most widely used.
Application of immune repertoire sequencing
(1)Application of immune repertoire sequencing in diagnosis of hematological tumors
Hematological tumors are caused by mutations in genes. On the basis of different phenotypes, known or unknown mutations occur in genes, thus causing the occurrence of hematological tumors. However, deep sequencing of CDR3 gene sequence of B or T lymphocytes using high-throughput sequencing technology can diagnose hematological tumors.
(2)Application of immune repertoire sequencing in evaluating immune reconstitution after bone marrow transplantation
Hematopoietic stem cell transplantation is the most effective treatment for hematological tumors. On the one hand, transplanted bone marrow hematopoietic stem cells can rebuild immune function to resist abnormal tumor cells; on the other hand, transplanted T cells can recognize tumor-related antigens and remove these abnormal tumor cells. In recent years, the use of TCR or immunoglobulin gene sequencing studies can be used to assess the status of T cell and B cell immune reconstitution after hematopoietic stem cell transplantation.
(3)Application of immune repertoire sequencing in the monitoring of minimal residual lesions
In acute lymphoblastic leukemia, minimal residual lesions are recognized as the best independent prognostic factor, and initially defined as minimal residual lesions to assess therapeutic efficacy. They are later used to monitor early recurrence of tumors. Current surveillance methods for acute lymphoblastic leukemia include the monitoring of abnormal immune phenotypes of leukemia by flow immunoassay. Although these methods have proved feasible in clinical practice, the monitoring of different leukemia subclones during leukemia treatment by flow immunoassay and multiplex polymerase chain reaction has some limitations or cannot be detected, which may lead to false results. The monitoring of clonal immunoglobulin or TCR rearrangement gene sequences by high-throughput immunohistology library sequencing technology is helpful for the diagnosis of minimal residual lesions in acute lymphoblastic leukemia.
(3)Application of immune repertoire sequencing in evaluation of the efficacy of new tumor drugs and exploration of mechanisms
The emergence of immune checkpoint inhibitors has brought new hope to cancer patients, from which some patients with relapse/refractory disease can benefit, and achieve even long-term disease-free survival. The population of patients who are suitable for treatment with immune checkpoint inhibitors and their specific mechanisms are not known at present, but immune checkpoint inhibitors may affect and change the ability of the immune system to fight tumors to some extent. Therefore, the detection of immune repertoire before and after treatment may become an effective method to further explore the mechanism of action of this drug and to find immunological markers to predict the efficacy.
From traditional flow cytometry, immune scanning lineage analysis technology to high-throughput sequencing technology, the sequencing technology of immune repertoire has undergone revolutionary development. With the continuous progress of amplification technology, sequencing technology and data analysis technology, the sequencing technology of immune repertoire will play an increasingly important role in the early diagnosis, treatment and evaluation of hematological tumors. Through TCR and BCR receptor bank analysis, interpreting the tumorigenesis and development at the DNA molecular level, will bring new hope for the evaluation of tumor recurrence, minimal residual disease, and immune reconstitution after hematopoietic stem cell transplantation. However, due to the complexity of BCR and TCR rearrangements and the diversity of CD receptor libraries, the depth of high-throughput sequencing, and the interpretation and application of huge data, there will be many difficulties, so the immune repertoire technology is currently less applied and has not been used for clinical diagnosis and treatment of diseases.